海蛇尾环境DNA宏条形码技术的两套PCR引物构建
Development of two new sets of PCR primers for eDNA metabarcoding of brittle stars
作者:Masanori Okanishi, Hisanori Kohtsuka, Qianqian Wu, Junpei Shinji, Naoki Shibata, Takashi Tamada, Tomoyuki Nakano, Toshifumi Minamoto
期刊:Metabarcoding and Metagenomics
Brittle stars (class Ophiuroidea) are marine invertebrates comprising approximately 2,100 extant species, and are considered to constitute the most diverse taxon of the phylum Echinodermata. As a non-invasive method for monitoring biodiversity, we developed two new sets of PCR primers for metabarcoding environmental DNA (eDNA) from brittle stars. The new primer sets were designed to amplify 2 short regions of the mitochondrial 16S rRNA gene, comprising a conserved region (111–115 bp, 112 bp on average; named “16SOph1”) and a hyper-variable region (180195 bp, 185 bp on average; named “16SOph2”) displaying interspecific variation. The performance of the primers was tested using eDNA obtained from two sources: a) rearing water of an 2.5 or 170 L aquarium tanks containing 15 brittle star species and b) from natural seawater collected around Misaki, the Pacific coast of central Japan, at depths ranging from shallow (2 m) to deep (> 200 m) sea. To build a reference library, we obtained 16S rRNA sequences of brittle star specimens collected from around Misaki and from similar depths in Japan, and sequences registered in International Nucleotide Sequence Database Collaboration. As a result of comparison of the obtained eDNA sequences with the reference library 37 (including cryptic species) and 26 brittle star species were detected with certain identities by 16SOph1 and 16SOph2 analyses, respectively. In shallow water, the number of species and reads other than the brittle stars detected with 16SOph1 was less than 10% of the total number. On the other hand, the number of brittle star species and reads detected with 16SOph2 was less than half of the total number, and the number of detected non-brittle star metazoan species ranged from 20 to 46 species across 6 to 8 phyla (only the reads at the “Tank” were less than 0.001%). The number of non-brittle star species and reads at 80 m was less than 10% with both of the primer sets. These findings suggest that 16SOph1 is specific to the brittle star and 16SOph2 is suitable for a variety of marine metazoans. It appears, however, that further optimization of primer sequences would still be necessary to avoid possible PCR dropouts from eDNA extracts. Moreover, a detailed elucidation of the brittle star fauna in the examined area, and the accurate identification of brittle star species in the current DNA databank is required.
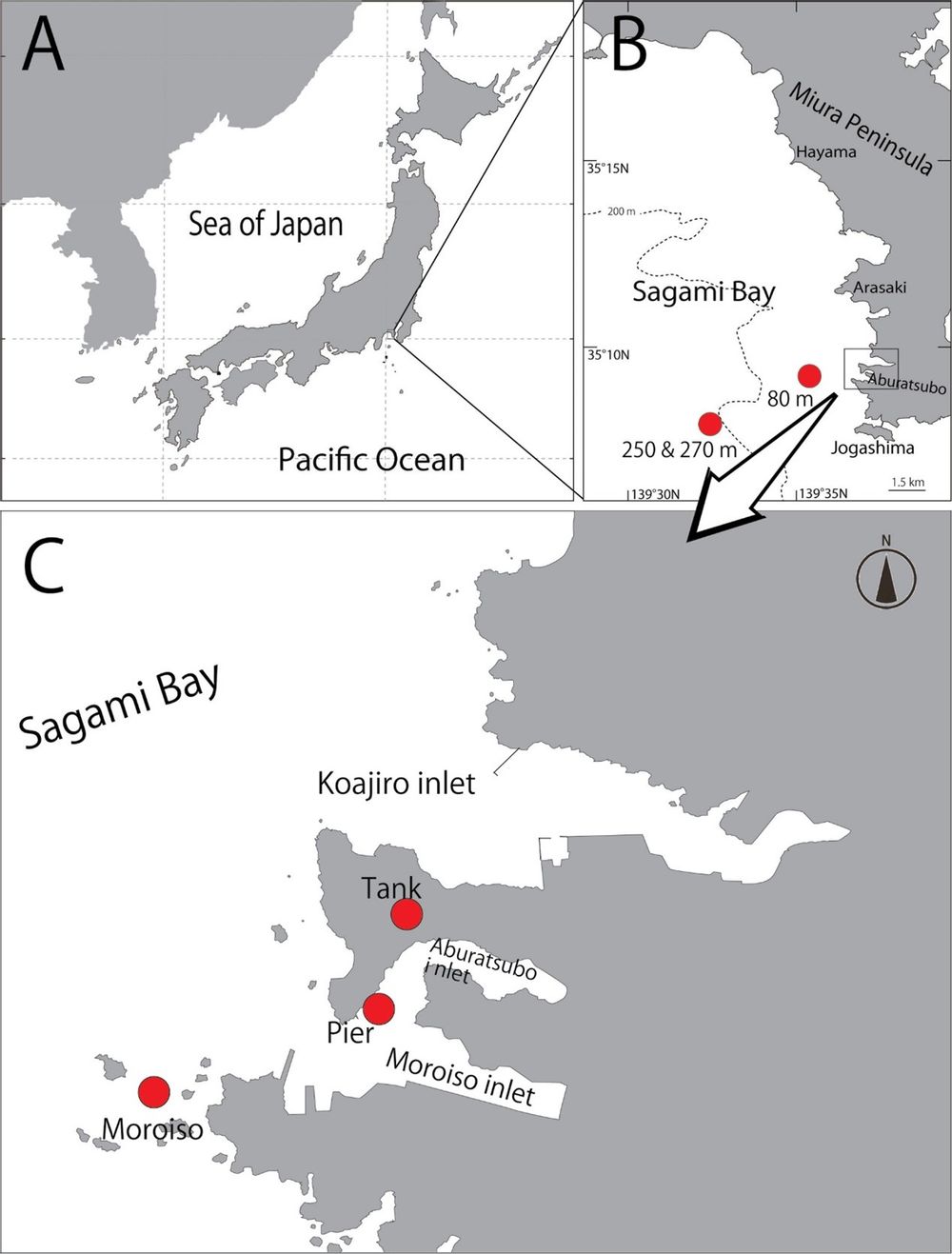
Figure 1. A–C. Map of Japan (A), showing the location of the 2 sea water sampling sites of Misaki Marina Biological Station (B) and 2 sea water sampling sites at shallow waters and aquarium tank (C) (Koajiro, Misaki, Miura, Kanagawa Prefecture).
海蛇尾(蛇尾纲)是一种海洋无脊椎动物,现存约2100个物种,被认为是棘皮动物门中最多样化的分类单元。作为一种用于监测生物多样性的非侵入性方法,本研究开发了两套全新的用于海蛇尾宏条形码技术环境DNA(eDNA)的聚合酶链式反应(PCR)引物。新的引物用于扩增线粒体16S rRNA基因的两个短区域,包括一个保守区(111-115个碱基对,平均112个碱基对;命名为“16SOph1”)和一个显示种间变异的高变区(180-195个碱基对,平均185个碱基对;命名为“16SOph2”)。引物的性能分别使用以下两个来源获得的eDNA进行测试:a)含有15种海蛇尾物种的2.5升或170升的水族箱饲养水,和b)从日本中部太平洋海岸三崎附近收集的天然海水,深度为从浅海(2米)至深海(大于200米)。为了建立一个参考数据库,本研究获取了从三崎周围及类似深度日本海域采集的海蛇尾标本的16S rRNA序列,和在国际协作核酸序列数据库中已登记的序列。将所获得的eDNA序列与参考数据库进行比较,通过16SOph1和16SOph2分析,分别检测到37个(包括隐存种)和26个具有一定特征的海蛇尾物种。在浅水区,用16SOph1检测到的非海蛇尾的物种数量和测序序列数量不到总数的10%。用16SOph2检测到的海蛇尾物种数量和测序序列数量不到总数的一半,检测到的非海蛇尾后生动物物种数量为20种到46种不等,涉及6到8个门类(只有水族箱样本的测序序列少于0.001%)。在取自海水深度为80米的天然海水样本中,两套引物测出来的的非海蛇尾物种数和测序序列均少于10%。这些研究结果表明,16SOph1对海蛇尾具有特异性,而16SOph2则适用于各种海洋后生动物。然而,为了避免eDNA提取物中可能会出现的PCR缺失,仍然有必要进一步优化引物序列。此外,还需要详细阐明研究区域的海蛇尾动物群,并在现有DNA数据库中准确识别海蛇尾物种。








